Writing Reports with R Markdown
Overview
Teaching: 75 min
Exercises: 15 minQuestions
How can I make reproducible reports using R Markdown?
How do I format text using Markdown?
Objectives
To create a report in R Markdown that combines text, code, and figures.
To use Markdown to format our report.
To understand how to use R code chunks to include or hide code, figures, and messages.
To be aware of the various report formats that can be rendered using R Markdown.
Contents
- R for data analysis review
- What is R Markdown and why use it?
- Creating an R Markdown file
- Basic components of R Markdown
- Starting the report
- Customizing the report
- Formatting
- Applying it to your own data
R for data analysis review
Review: Creating summaries
Read in the 1990 smoking dataset and find the mean, median, min, and max population for each continent.
Solution
library(tidyverse)Warning: package 'ggplot2' was built under R version 4.3.3Warning: package 'tibble' was built under R version 4.3.3Warning: package 'purrr' was built under R version 4.3.3Warning: package 'lubridate' was built under R version 4.3.3── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ── ✔ dplyr 1.1.4 ✔ readr 2.1.5 ✔ forcats 1.0.0 ✔ stringr 1.5.1 ✔ ggplot2 3.5.2 ✔ tibble 3.3.0 ✔ lubridate 1.9.4 ✔ tidyr 1.3.1 ✔ purrr 1.0.4 ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ── ✖ dplyr::filter() masks stats::filter() ✖ dplyr::lag() masks stats::lag() ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorssmoking <- read_csv('data/smoking_cancer_1990.csv')Rows: 191 Columns: 6 ── Column specification ──────────────────────────────────────────────────────── Delimiter: "," chr (2): country, continent dbl (4): year, pop, smoke_pct, lung_cancer_pct ℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.smoking %>% group_by(continent) %>% summarise(mean_pop = mean(pop), median_pop = median(pop), min_pop = min(pop), max_pop = max(pop))# A tibble: 6 × 5 continent mean_pop median_pop min_pop max_pop <chr> <dbl> <dbl> <dbl> <dbl> 1 Africa 11655968. 6788686. 69507 95212454 2 Asia 75684706. 12446168 223159 1135185000 3 Europe 13056650. 5140939 24124 79433029 4 North America 18257642. 2470946 40260 249623000 5 Oceania 1907505 121440. 8910 17065100 6 South America 24606701. 11752774 405169 149003225
What is R Markdown and why use it?
Recall that our goal is to generate a report on how a country’s smoking rate is related to its lung cancer rate.
Discussion
How do you usually share data analyses with your collaborators?
Solution
Many people share them through a Word or PDF document, a spreadsheet, slides, a graphic, etc.
In R Markdown, you can incorporate ordinary text (ex. experimental methods, analysis and discussion of results) alongside code and figures! (Some people write entire manuscripts in R Markdown.) This is useful for writing reproducible reports and publications, sharing work with collaborators, writing up homework, and keeping a bioinformatics notebook. Because the code is emedded in the document, the tables and figures are reproducible. Anyone can run the code and get the same results. If you find an error or want to add more to the report, you can just re-run the document and you’ll have updated tables and figures! This concept of combining text and code is called “literate programming”. To do this we use R Markdown, which combines Markdown (renders plain text) with R. You can output an html, PDF, or Word document that you can share with others. In fact, this webpage is an example of a rendered R markdown file!
(If you are familiar with Jupyter notebooks in the Python programming environment, R Markdown is R’s equivalent of a Jupyter notebook.)
Creating an R Markdown file
Now that we have a better understanding of what we can use R Markdown files for, let’s start writing a report!
To create an R Markdown file:
- Open RStudio
- Go to File → New File → R Markdown
- Give your document a title, something like “The relationship between smoking and lung cancer rates” (Note: this is not the same as the file name - it’s just a title that will appear at the top of your report)
- Keep the default output format as HTML
- Note: R Markdown files always end in
.Rmd
R Markdown Outputs
The default output for an R Markdown report is HTML, but you can also use R Markdown to output other report formats. For example, you can generate PDF reports using R Markdown, but you must install TeX to do this.
Next, let’s save our R markdown file to the reports directory.
You can do this by clicking the save icon in the top left or using control + s (command + s on a Mac).
Change knit directory
There’s one other thing that we need to do before we get started with our report.
To render our documents into html format, we can “knit” them in R Studio.
Usually, R Markdown renders documents from the directory where the document is saved (the location of the .Rmd file), but we want it to render from the main project directory where our .Rproj file is.
This is because that’s where all of our relative paths are from and it’s good practice to have all of your relative paths from the main project directory.
To change this default, click on the down arrow next to the “Knit” button at the top left of R Studio, go to “Knit Directory” and click “Project Directory”.
Now it will assume all of your relative paths for reading and writing files are from the un-report directory, rather than the reports directory.
Now that we have that set up, let’s start on the report!
Add code
We’re going to use the code you generated yesterday to plot smoking rates vs. lung cancer rates to include in the report. Recall that we needed a couple R packages to generate these plots. We can create a new code chunk to load the needed packages. You could also include this in the previous setup chunk, it’s up to your personal preference. To create a new code chunk, you have several options: type it out yourself, click the button with the green c and + in the top right, next to Run, or use the keyboard shortcut Ctrl + Alt + i.
```{r packages}
library(tidyverse)
```
Now, in a real report this is when we would type out the background and purpose of our analysis to provide context to our readers. However, since writing is not a focus of this workshop we will avoid lengthy prose and stick to short descriptions. You can copy the following text into your own report below the package code chunk.
This report was prepared to analyze the relationship between a country's lung cancer rate, smoking rate, and air pollution. Our goal is to determine to what degree the percent of people who smoke and the amount of air pollution per capita may be related to its lung cancer rate. We hypothesize that lung cancer rates increase with both percent of people who smoke and the amount of air pollution per capita.
Now, since we want to show our results comparing smoking rate and lung cancer rate by country, we need to read in this data so we can generate our plot. We will add another code chunk to prepare the data.
```{r data}
smoking <- read_csv("data/smoking_cancer.csv")
```
Plot
Now that we have the data, we need to produce the plot. Let’s create it using the most recent year in our dataset:
```{r smoking_cancer}
smoking %>%
filter(year == max(year)) %>%
ggplot() +
aes(x = smoke_pct, y = lung_cancer_pct, color=continent, size=pop/1000000) +
geom_point() +
labs(x = "Percent of people who smoke", y = "Percent of people with lung cancer",
title= "Are lung cancer rates associated with smoking rates?", size="Population (in millions)")
```
Table
Let’s say we also want to include a table in our report that summarizes the the minimum, median, and maximum smoker percent.
```{r}
smoking %>%
summarize(min_smoke = min(smoke_pct),
median_smoke = median(smoke_pct),
max_smoke = max(smoke_pct))
```
Knitting
Now we can knit our document to see how our report looks! Use the knit button in the top left of the screen.

Amazing! We’ve created a report!
It’s looking pretty good, but there seem to be a few extra bits that we might not need in the report. For example, what if we want to make a report that doesn’t print out all of the messages from the tidyverse? Or a report that doesn’t show the code? And the table is a bit ugly. Let’s make things a bit prettier.
Customizing the report
Table format
We can make the table prettier using the R function kable(). We can give the kable() function a tibble and it will format it to a nice looking table in the report. The kable() function comes from the knitr packages, so what do we have to do before using the function? We have to load the knitr library.
# load library
library(knitr)
# print kable
smoking %>%
summarize(min_smoke = min(smoke_pct),
median_smoke = median(smoke_pct),
max_smoke = max(smoke_pct)) %>%
kable()
| min_smoke| median_smoke| max_smoke|
|---------:|------------:|---------:|
| 3.118639| 24.36522| 46.91547|
Messages
How do we get rid of the tidyverse messages? One way to do this is by saying include = FALSE in the curly brackets for that code chunk:
```{r packages, include=FALSE}
library(tidyverse)
```
This will get rid of the code and the corresponding messages. But what if we want to include the code so that people know we loaded the tidyverse? In this case, we can say message=FALSE inside the curly brackets:
```{r packages, message=FALSE}
library(tidyverse)
```
Code
We can also see the code that was used to generate the plot. Depending on the purpose and audience for your report, you may or may not want to include the code. If you don’t want the code to appear, how can you prevent it?
Code chunk options
Which of the following would lead to a report with no code chunks? For the ones that would not work, what would happen instead?
- Add
echo = FALSEinside the curly brackets of the plotting code chunk.- Add
include = FALSEinside the curly brackets of the plotting code chunk.- Add
echo = FALSEinside the curly brackets of the setup code chunk.- Change
echo = TRUEtoecho = FALSEin theknitr::opts_chunk$set()function in the first code chunk.Solution
- This would only remove the code for the plotting code chunk, but not the packages code chunk.
- This would remove the code but also the plot from the report.
- This would not change anything because that code chunk is already excluded because
include = FALSE.- This would remove all code from the output file (what we want).
Add to our report: association between air pollution and lung cancer
We have a pretty nice looking report, but we still haven’t included anything about the association between lung cancer and air pollution per capita. Let’s add a section to our markdown document in the following steps:
- Make a new header and write a 1-2 sentence description of what you will be plotting.
- Create a new code chunk
- Make a plot with air pollution per capita on the x axis and lung cancer on the y axis
- Make a table with summary statistics including the minimum, median, and maximum air pollution values.
- BONUS: Merge the table we created earlier with the table you created here with rows for smoking and air pollution, and a column for each of the summary statistics.
Solution
One option to create a code chunk is to type it out. You can also see other options above. Then you have to read in the data and create the plot and table:
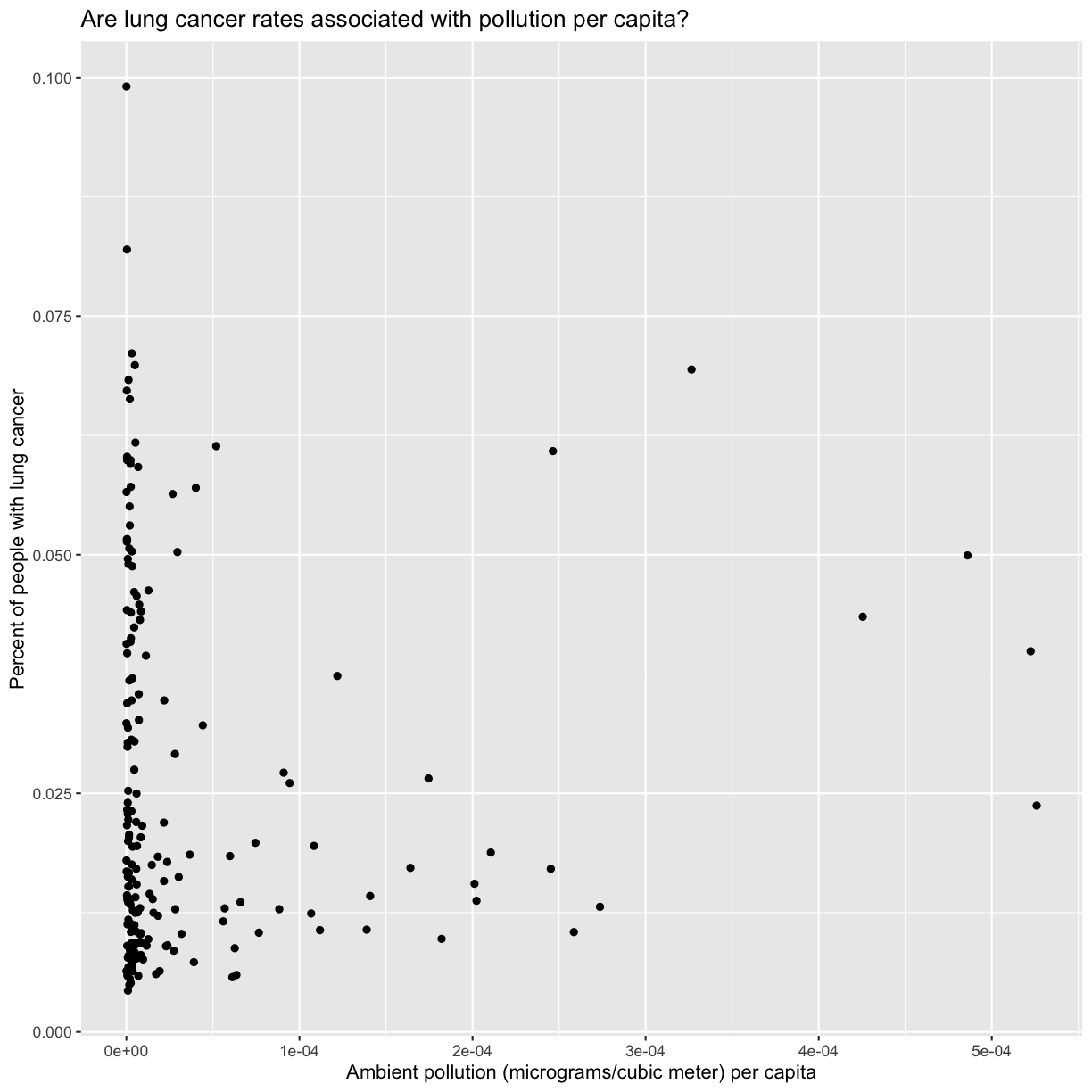
smoking_pollution <- read_csv('data/smoking_pollution.csv')Rows: 191 Columns: 7 ── Column specification ──────────────────────────────────────────────────────── Delimiter: "," chr (2): country, continent dbl (5): year, pop, smoke_pct, lung_cancer_pct, pollution ℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.smoking_pollution %>% ggplot(aes(x = pollution/pop, y = lung_cancer_pct)) + geom_point() + labs(x = "Ambient pollution (micrograms/cubic meter) per capita", y = "Percent of people with lung cancer", title = "Are lung cancer rates associated with pollution per capita?")
smoking_pollution %>% summarize(min_pol = min(pollution), median_smoke = median(pollution), max_smoke = max(pollution))# A tibble: 1 × 3 min_pol median_smoke max_smoke <dbl> <dbl> <dbl> 1 4.69 24.5 78.2Bonus: you can use
pivot_longer()andgroup_by()followed bysummarize():smoking_pollution %>% pivot_longer(c(smoke_pct, pollution)) %>% group_by(name) %>% summarize(min = min(value), median = median(value), max = max(value))# A tibble: 2 × 4 name min median max <chr> <dbl> <dbl> <dbl> 1 pollution 4.69 24.5 78.2 2 smoke_pct 3.12 24.4 46.9Notice that we used
c()to providepivot_longer()with the two column names that we wanted to pivot.c()stands for “combine”; this function combines the two values into what we call a vector.
Bonus: Dynamically updating text using mini R chunks
Sometimes, you want to describe your data or results (like our plot) to the audience in text but the data and results may still change as you work things out. R Markdown offers an easy way to do this dynamically, so that the text updates as your data or results change!
Say we want to get the number of countries present in our dataset. Previously, we learned about the
distinct()function that returns distinct values. There’s a very similar function calledn_distinct()that returns instead the number of distinct values:n_countries <- smoking %>% select(country) %>% n_distinct()Now, all we need to do is reference the values we just computed to describe our plot. To do this, we enclose each value in one set of backticks (
`r some_R_variable_name `), while therpart once again indicates that it’s a chunk of R code. When we knit our report, R will automatically fill in the values we just created in the above code chunk. Note that R will automatically update these values every time our data might change (if we were to decide to drop or add countries to this analysis, for example).There are `r n_countries ` countries in our dataset.Try knitting the document and see what happens!
Key Points
R Markdown is a useful way to create a report that integrates text, code, and figures.
Options such as
includeandechodetermine what parts of an R code chunk are included in the R Markdown report.R Markdown can render HTML, PDF, and Microsoft Word outputs.